Update Meeting: 11-11-2021
Jenny Sjaarda
2021-11-11
Last updated: 2021-11-22
Checks: 7 0
Knit directory: proxyMR/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20210602) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 810aae7. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: _targets/
Ignored: analysis/_site.yml_cp
Ignored: analysis/bgenie_GWAS/
Ignored: analysis/data_setup/
Ignored: analysis/download_Neale_list.csv
Ignored: analysis/process_Neale.out
Ignored: analysis/traitMR/
Ignored: data/Neale_SGG_directory_12_07_2021.csv
Ignored: data/Neale_SGG_directory_15_07_2021.csv
Ignored: data/PHESANT_file_directory_05_10_2021.txt
Ignored: data/UKBB_pheno_directory_05_10_2021.csv
Ignored: data/processed/
Ignored: data/raw/
Ignored: output/figures/
Ignored: output/tables/traitMR/
Ignored: proxyMR_comparison.RData
Ignored: proxyMR_figure_data.RData
Ignored: proxymr_100_clustermq.out
Ignored: proxymr_101_clustermq.out
Ignored: proxymr_102_clustermq.out
Ignored: proxymr_103_clustermq.out
Ignored: proxymr_104_clustermq.out
Ignored: proxymr_105_clustermq.out
Ignored: proxymr_106_clustermq.out
Ignored: proxymr_107_clustermq.out
Ignored: proxymr_108_clustermq.out
Ignored: proxymr_109_clustermq.out
Ignored: proxymr_10_clustermq.out
Ignored: proxymr_110_clustermq.out
Ignored: proxymr_111_clustermq.out
Ignored: proxymr_112_clustermq.out
Ignored: proxymr_113_clustermq.out
Ignored: proxymr_114_clustermq.out
Ignored: proxymr_115_clustermq.out
Ignored: proxymr_116_clustermq.out
Ignored: proxymr_117_clustermq.out
Ignored: proxymr_118_clustermq.out
Ignored: proxymr_119_clustermq.out
Ignored: proxymr_11_clustermq.out
Ignored: proxymr_120_clustermq.out
Ignored: proxymr_121_clustermq.out
Ignored: proxymr_122_clustermq.out
Ignored: proxymr_123_clustermq.out
Ignored: proxymr_124_clustermq.out
Ignored: proxymr_125_clustermq.out
Ignored: proxymr_126_clustermq.out
Ignored: proxymr_127_clustermq.out
Ignored: proxymr_128_clustermq.out
Ignored: proxymr_129_clustermq.out
Ignored: proxymr_12_clustermq.out
Ignored: proxymr_130_clustermq.out
Ignored: proxymr_131_clustermq.out
Ignored: proxymr_132_clustermq.out
Ignored: proxymr_133_clustermq.out
Ignored: proxymr_134_clustermq.out
Ignored: proxymr_135_clustermq.out
Ignored: proxymr_136_clustermq.out
Ignored: proxymr_137_clustermq.out
Ignored: proxymr_138_clustermq.out
Ignored: proxymr_139_clustermq.out
Ignored: proxymr_13_clustermq.out
Ignored: proxymr_140_clustermq.out
Ignored: proxymr_14_clustermq.out
Ignored: proxymr_15_clustermq.out
Ignored: proxymr_16_clustermq.out
Ignored: proxymr_17_clustermq.out
Ignored: proxymr_18_clustermq.out
Ignored: proxymr_19_clustermq.out
Ignored: proxymr_1_clustermq.out
Ignored: proxymr_20_clustermq.out
Ignored: proxymr_21_clustermq.out
Ignored: proxymr_22_clustermq.out
Ignored: proxymr_23_clustermq.out
Ignored: proxymr_24_clustermq.out
Ignored: proxymr_25_clustermq.out
Ignored: proxymr_26_clustermq.out
Ignored: proxymr_27_clustermq.out
Ignored: proxymr_28_clustermq.out
Ignored: proxymr_29_clustermq.out
Ignored: proxymr_2_clustermq.out
Ignored: proxymr_30_clustermq.out
Ignored: proxymr_31_clustermq.out
Ignored: proxymr_32_clustermq.out
Ignored: proxymr_33_clustermq.out
Ignored: proxymr_34_clustermq.out
Ignored: proxymr_35_clustermq.out
Ignored: proxymr_36_clustermq.out
Ignored: proxymr_37_clustermq.out
Ignored: proxymr_38_clustermq.out
Ignored: proxymr_39_clustermq.out
Ignored: proxymr_3_clustermq.out
Ignored: proxymr_40_clustermq.out
Ignored: proxymr_41_clustermq.out
Ignored: proxymr_42_clustermq.out
Ignored: proxymr_43_clustermq.out
Ignored: proxymr_44_clustermq.out
Ignored: proxymr_45_clustermq.out
Ignored: proxymr_46_clustermq.out
Ignored: proxymr_47_clustermq.out
Ignored: proxymr_48_clustermq.out
Ignored: proxymr_49_clustermq.out
Ignored: proxymr_4_clustermq.out
Ignored: proxymr_50_clustermq.out
Ignored: proxymr_51_clustermq.out
Ignored: proxymr_52_clustermq.out
Ignored: proxymr_53_clustermq.out
Ignored: proxymr_54_clustermq.out
Ignored: proxymr_55_clustermq.out
Ignored: proxymr_56_clustermq.out
Ignored: proxymr_57_clustermq.out
Ignored: proxymr_58_clustermq.out
Ignored: proxymr_59_clustermq.out
Ignored: proxymr_5_clustermq.out
Ignored: proxymr_60_clustermq.out
Ignored: proxymr_61_clustermq.out
Ignored: proxymr_62_clustermq.out
Ignored: proxymr_63_clustermq.out
Ignored: proxymr_64_clustermq.out
Ignored: proxymr_65_clustermq.out
Ignored: proxymr_66_clustermq.out
Ignored: proxymr_67_clustermq.out
Ignored: proxymr_68_clustermq.out
Ignored: proxymr_69_clustermq.out
Ignored: proxymr_6_clustermq.out
Ignored: proxymr_70_clustermq.out
Ignored: proxymr_71_clustermq.out
Ignored: proxymr_72_clustermq.out
Ignored: proxymr_73_clustermq.out
Ignored: proxymr_74_clustermq.out
Ignored: proxymr_75_clustermq.out
Ignored: proxymr_76_clustermq.out
Ignored: proxymr_77_clustermq.out
Ignored: proxymr_78_clustermq.out
Ignored: proxymr_79_clustermq.out
Ignored: proxymr_7_clustermq.out
Ignored: proxymr_80_clustermq.out
Ignored: proxymr_81_clustermq.out
Ignored: proxymr_82_clustermq.out
Ignored: proxymr_83_clustermq.out
Ignored: proxymr_84_clustermq.out
Ignored: proxymr_85_clustermq.out
Ignored: proxymr_86_clustermq.out
Ignored: proxymr_87_clustermq.out
Ignored: proxymr_88_clustermq.out
Ignored: proxymr_89_clustermq.out
Ignored: proxymr_8_clustermq.out
Ignored: proxymr_90_clustermq.out
Ignored: proxymr_91_clustermq.out
Ignored: proxymr_92_clustermq.out
Ignored: proxymr_93_clustermq.out
Ignored: proxymr_94_clustermq.out
Ignored: proxymr_95_clustermq.out
Ignored: proxymr_96_clustermq.out
Ignored: proxymr_97_clustermq.out
Ignored: proxymr_98_clustermq.out
Ignored: proxymr_99_clustermq.out
Ignored: proxymr_9_clustermq.out
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: Rplots.pdf
Unstaged changes:
Modified: analysis/AM_MR_summary.Rmd
Modified: analysis/couple_selection.Rmd
Modified: output/tables/define_Neale_categories.csv
Modified: output/tables/define_Neale_categories_filled.csv
Modified: renv.lock
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/update_meeting_11_11_2021.Rmd) and HTML (docs/update_meeting_11_11_2021.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 810aae7 | Jenny Sjaarda | 2021-11-22 | wflow_publish("analysis/update_meeting_11_11_2021.Rmd") |
| Rmd | 9845a87 | jennysjaarda | 2021-11-11 | add update meeting 11-11-21 |
| html | 9845a87 | jennysjaarda | 2021-11-11 | add update meeting 11-11-21 |
1 Progress update.
- Last time, we ran a MVMR as follows: \(Y_p \sim X_i + Y_i + X_p\), using instruments for \({X_i, Y_i, X_p}\). Unfortunately, realized this won't work after all because there is too much co-linearity between \(X_i\) and \(X_p\). We also don't have additional instruments for \(X_p\), beyond those that are already instruments for \(X_i\). The only way around would be to run a GWAS on \(X_p\), but probably not worth that effort (unlikely to find any additional instrument anyways).
- Updated pipeline to meta-analyze across sexes at the SNP level rather than the MR level (minimize weak-instrument bias).
- In all two-trait MRs, I’ve filtered out SNPs that show evidence of reverse causation based on same-person statistics (i.e. from Neale, meta-analyzed across sexes as per update above) and then use those same, filtered IVs in the couple MRs. IVs were filtered based on the code below (for reference), essentially removing any SNP that is more strongly associated with the outcome than the expsoure at a threshold of p <
# r reverse_MR_threshold, corresponding to thereverse_MR_thresholdin the code below.
reverse_t_threshold = stats::qnorm(reverse_MR_threshold)
dat_filter <- dat %>%
mutate(z_score.exposure = beta.exposure / se.exposure) %>%
mutate(z_score.outcome = beta.outcome / se.outcome) %>%
mutate(std_beta.exposure = z_score.exposure / sqrt(samplesize.exposure)) %>%
mutate(std_beta.outcome = z_score.outcome / sqrt(samplesize.outcome)) %>%
mutate(std_se.exposure = 1 / sqrt(samplesize.exposure)) %>%
mutate(std_se.outcome = 1 / sqrt(samplesize.outcome)) %>%
mutate(std_t_stat = (abs(std_beta.exposure) - abs(std_beta.outcome)) /
sqrt(std_se.exposure^2 + std_se.outcome^2)) %>%
filter(std_t_stat > reverse_t_threshold)2 Results.
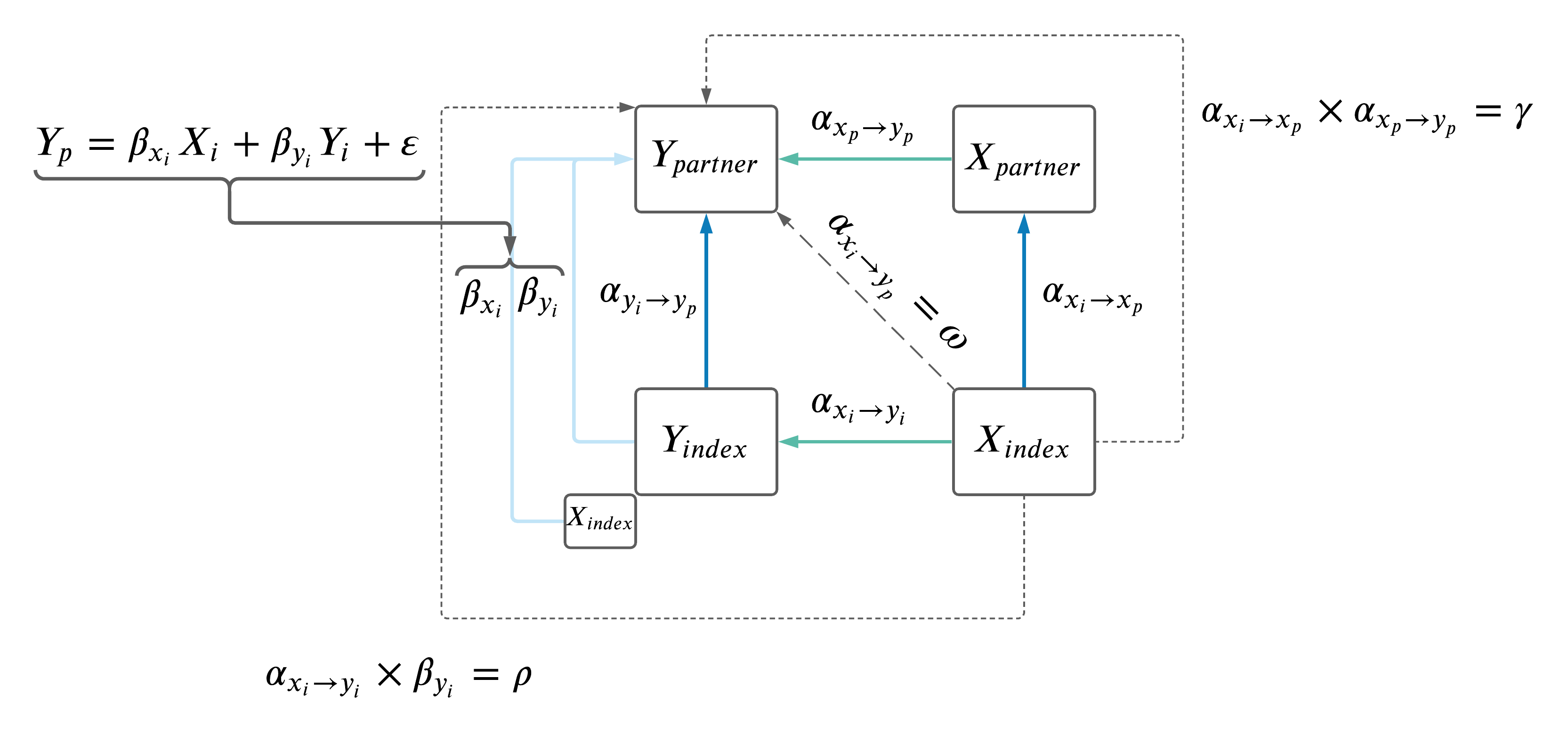
As a recap, the model is below.
| Version | Author | Date |
|---|---|---|
| 529020f | jennysjaarda | 2021-09-24 |
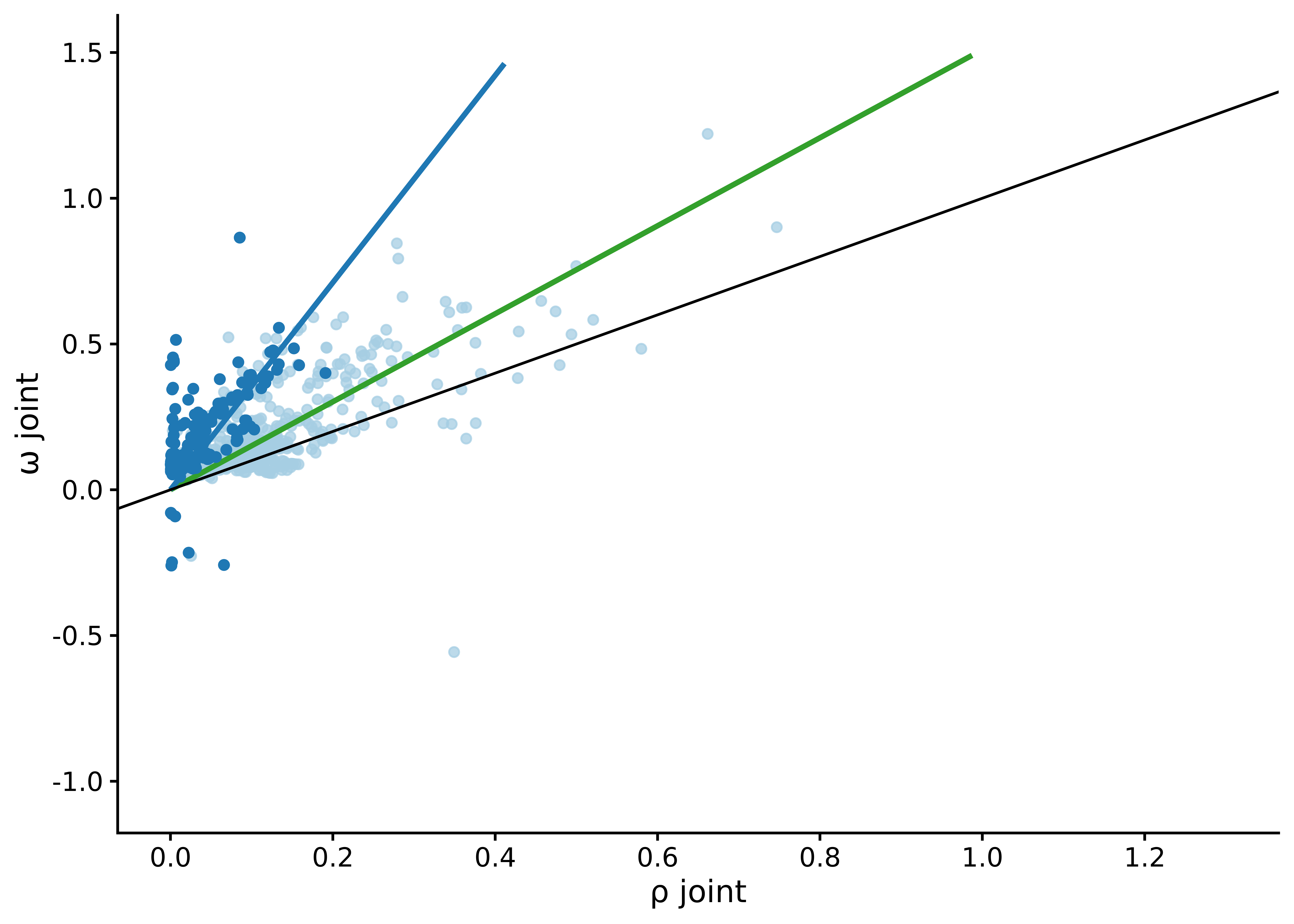
A summary of the regressions between \(\omega\), \(\rho\) and \(\gamma\) is below.
Call:
lm(formula = rho_beta ~ omega_beta + 0, data = data)
Residuals:
Min 1Q Median 3Q Max
-0.59720 -0.02468 0.00983 0.04595 0.36472
Coefficients:
Estimate Std. Error t value Pr(>|t|)
omega_beta 0.445129 0.009718 45.8 <0.0000000000000002 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.06695 on 929 degrees of freedom
Multiple R-squared: 0.6931, Adjusted R-squared: 0.6928
F-statistic: 2098 on 1 and 929 DF, p-value: < 0.00000000000000022
Call:
lm(formula = gam_beta ~ omega_beta + 0, data = data)
Residuals:
Min 1Q Median 3Q Max
-0.75402 -0.02179 0.01994 0.04937 1.10864
Coefficients:
Estimate Std. Error t value Pr(>|t|)
omega_beta 0.62689 0.01225 51.18 <0.0000000000000002 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.08438 on 929 degrees of freedom
Multiple R-squared: 0.7382, Adjusted R-squared: 0.7379
F-statistic: 2620 on 1 and 929 DF, p-value: < 0.00000000000000022
Call:
lm(formula = gam_rho_beta ~ omega_beta + 0, data = data)
Residuals:
Min 1Q Median 3Q Max
-1.18334 -0.02240 0.02567 0.07289 1.45454
Coefficients:
Estimate Std. Error t value Pr(>|t|)
omega_beta 1.07202 0.01664 64.42 <0.0000000000000002 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1147 on 929 degrees of freedom
Multiple R-squared: 0.8171, Adjusted R-squared: 0.8169
F-statistic: 4149 on 1 and 929 DF, p-value: < 0.00000000000000022
Call:
lm(formula = gam_rho_resid ~ omega_beta + 0, data = data)
Residuals:
Min 1Q Median 3Q Max
-0.76873 -0.02561 0.01144 0.04716 0.85106
Coefficients:
Estimate Std. Error t value Pr(>|t|)
omega_beta 0.72646 0.01171 62.02 <0.0000000000000002 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.0807 on 929 degrees of freedom
Multiple R-squared: 0.8054, Adjusted R-squared: 0.8052
F-statistic: 3846 on 1 and 929 DF, p-value: < 0.00000000000000022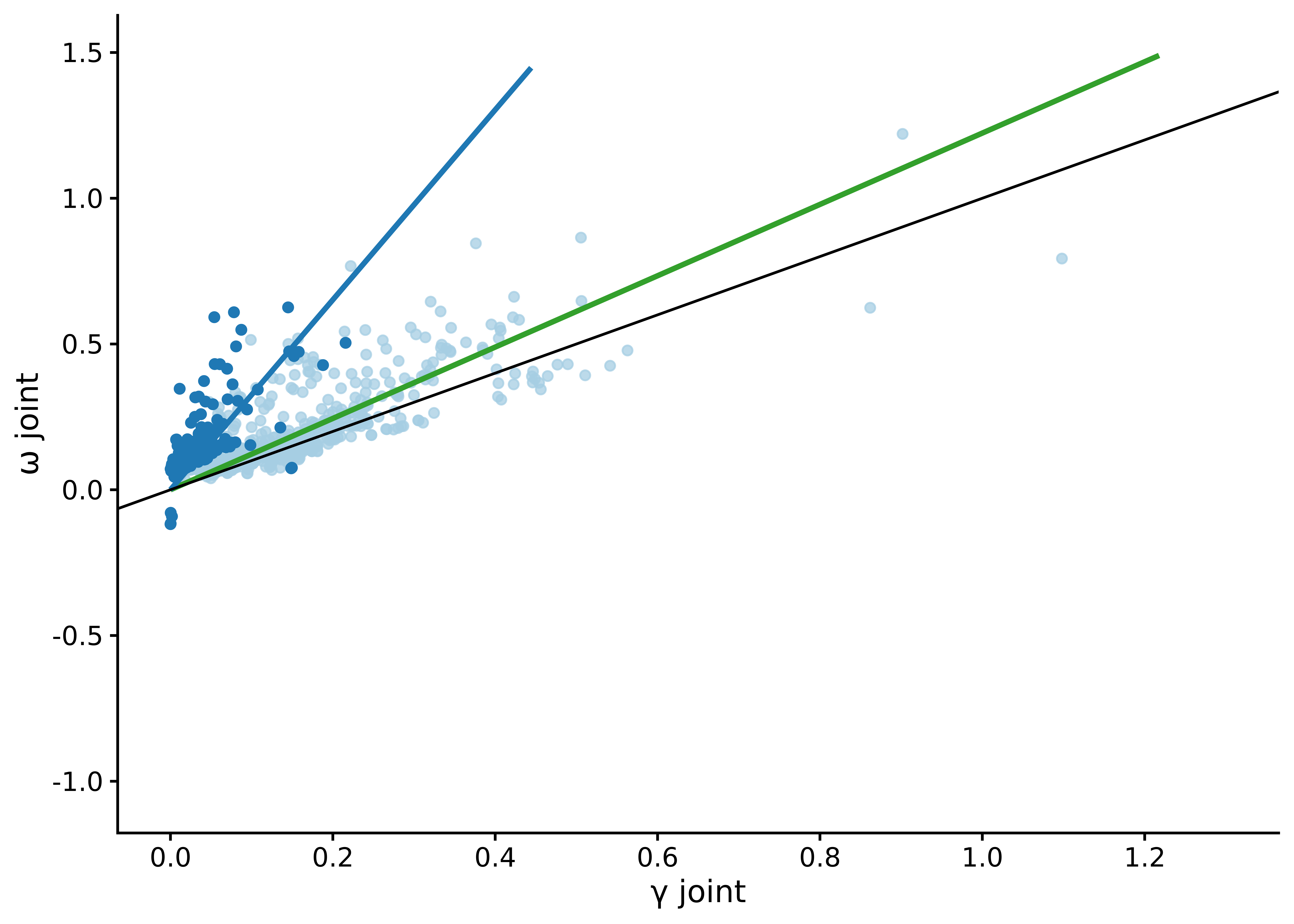
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /data/sgg2/jenny/bin/R-4.1.0/lib64/R/lib/libRblas.so
LAPACK: /data/sgg2/jenny/bin/R-4.1.0/lib64/R/lib/libRlapack.so
locale:
[1] en_CA.UTF-8
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] cowplot_1.1.1 kableExtra_1.3.4 knitr_1.33 DT_0.18.1
[5] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4
[9] readr_1.4.0 tidyr_1.1.3 tibble_3.1.2 ggplot2_3.3.4
[13] tidyverse_1.3.1 targets_0.5.0.9001 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.2 sass_0.4.0 splines_4.1.0 jsonlite_1.7.2
[5] viridisLite_0.4.0 modelr_0.1.8 bslib_0.3.0 assertthat_0.2.1
[9] highr_0.9 renv_0.13.2-62 cellranger_1.1.0 yaml_2.2.1
[13] lattice_0.20-44 pillar_1.6.1 backports_1.2.1 glue_1.4.2
[17] digest_0.6.27 promises_1.2.0.1 rvest_1.0.0 colorspace_2.0-1
[21] Matrix_1.3-3 htmltools_0.5.2 httpuv_1.6.1 pkgconfig_2.0.3
[25] broom_0.7.7 haven_2.4.1 scales_1.1.1 webshot_0.5.2
[29] processx_3.5.2 svglite_2.0.0 whisker_0.4 later_1.2.0
[33] git2r_0.28.0 mgcv_1.8-35 farver_2.1.0 generics_0.1.0
[37] ellipsis_0.3.2 withr_2.4.2 cli_2.5.0 magrittr_2.0.1
[41] crayon_1.4.1 readxl_1.3.1 evaluate_0.14 ps_1.6.0
[45] fs_1.5.0 fansi_0.5.0 nlme_3.1-152 xml2_1.3.2
[49] tools_4.1.0 data.table_1.14.0 hms_1.1.0 lifecycle_1.0.0
[53] munsell_0.5.0 reprex_2.0.0 callr_3.7.0 compiler_4.1.0
[57] jquerylib_0.1.4 systemfonts_1.0.2 rlang_0.4.11 grid_4.1.0
[61] rstudioapi_0.13 htmlwidgets_1.5.3 igraph_1.2.6 rmarkdown_2.11.2
[65] gtable_0.3.0 codetools_0.2-18 DBI_1.1.1 R6_2.5.0
[69] lubridate_1.7.10 fastmap_1.1.0 utf8_1.2.1 rprojroot_2.0.2
[73] stringi_1.6.2 Rcpp_1.0.6 vctrs_0.3.8 dbplyr_2.1.1
[77] tidyselect_1.1.1 xfun_0.24